Our analysis method
Microba’s advanced metagenomics (meaning many genomes) is used to analyse each sample and make sense of the community of microorganisms living in your gut microbiome. Microba’s powerful sequencing allows Insight™ to not only detect the many organisms in your sample but also to identify newly discovered species that may not be well known to science and see how they may be contributing to your overall health. By receiving this information, you will be able to see the whole picture not just part of it.
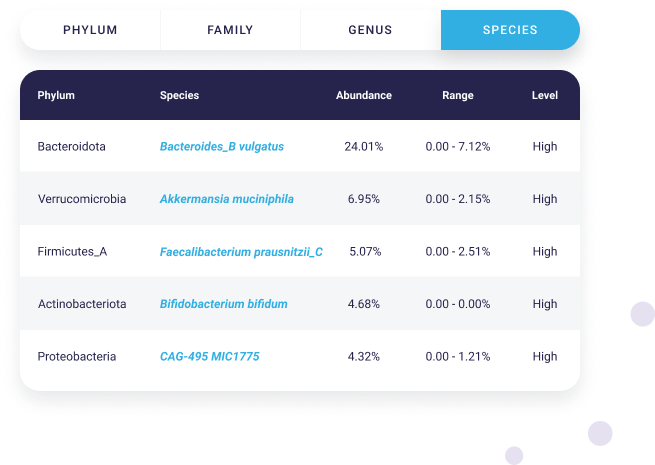
Get the specifics
We detect and identify thousands of microbial species, even at low levels, including Bacteroides, Akkermansia, Faecalibacterium, Bifidobacterium, Lactobacillus, Streptococcus, Escherichia, Clostridium, and many more!
See your microbiome from every taxonomic level, including phylum, family, genus, and species, as well as the varying degrees they are present.

Dive into the species
Looking for a specific species? Use the search function in your Microbiome Species Table to see if and at what level that microorganism appears within your microbiome. You can also download a CSV copy of your species list to save for later or to take to your next healthcare consultation.
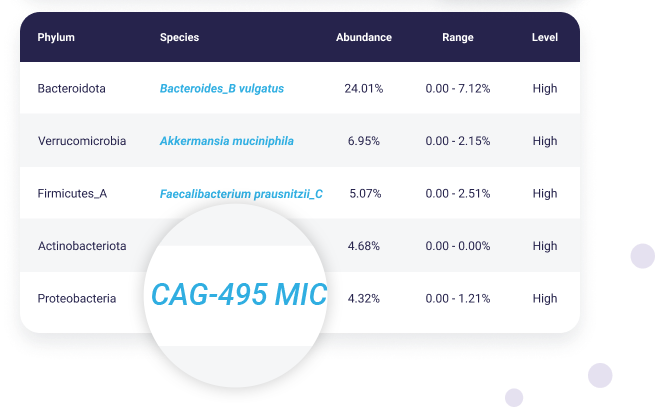
New discoveries
Newly discovered or less known species will appear in your report with code names, such as “CAG”, “UBA” or “MIC” and will have varying degrees of information attached to them.
Once all microorganisms’ DNA in your sample are sequenced and identified, Microba keeps a record of both known and newly discovered species in the Microba Genome Database. This reference database contains several tens of thousands of microorganisms, many of which do not exist in public databases. This enables Microba to identify at least 25% more DNA than other metagenomic classifiers and gives Insight™ access to a faster, more accurate approach than other analysis methods.
Microba’s method will detect most bacteria, archaea, fungi and parasites known to science, as well as identify new ones! If a species is included in our database, then we can detect that species if it is present above our detection limit in your sample.
Our growing database of microbial genomes uses the most scientifically advanced microbial taxonomy and complete database of bacterial and archaeal genomes currently available, known as the Genome Taxonomy Database.
Our own world-leading database is regularly updated with new genomes to improve species identification from samples. This is important because you may have newly identified species present in your gut performing functions that may be beneficial or detrimental to your overall health.
To check if a particular species you are interested in is detected by our test, you can check the Genome Taxonomy Database.
Please note, sometimes the names of bacterial species can change as more is learned about them and they are re-classified, so they may be present in the database under a different name.
Yes, our test will detect Helicobacter pylori (H. pylori) if it is present in your sample above our detection limit.
However, H. pylori is usually found in the stomach and because our test uses a stool sample for analysis, it can only detect the microorganisms that live in the lower part of your large intestine (colon). A stool sample is not representative of microorganisms that live higher up in the gastrointestinal tract, such as the stomach, small intestine or upper large intestine.
Therefore, you shouldn’t be surprised if your report doesn’t show H. pylori.
Yes, our test can detect Candida (Candida albicans), along with other yeasts and fungi. We report the organism’s relative abundance (the level of that organism as a percentage of the whole community).
If the proportion of Candida albicans in your sample is very low, it may not be detected since our method sequences all of the microorganisms in your sample instead of targeting specific species.
If you are primarily interested in detecting Candida and not the rest of the bacterial species present, we suggest using a Candida-specific test.

Our laboratory process
Microba’s state-of-the-art laboratory is located at Brisbane’s Translational Research Institute and uses Illumina's latest NovaSeq™ 6000 high throughput sequencing by Illumina Inc. Together, Microba's high quality gut microbiome analysis platform and Illumina's Next Generation Sequencing tools process thousands of samples and generate the most accurate metagenomic data available.
Meet the team